As you are probably aware, CUDA 7 was officially released during the 2015 GPU Technology Conference. For this Spotlight I took a few minutes to pick the brain of an early adopter of CUDA 7 to see how his work benefits from the new C++11 support.
I interviewed Yu-Hang Tang, a Ph.D. candidate in the Division of Applied Mathematics at Brown University in Providence, Rhode Island.
What breakthrough project is currently taking up all of your brain’s time?

At this moment we are finalizing a particle-based simulator for the in silico investigation of microfluidic devices used in cancer diagnostic. The code enables us to predict the behavior of cancer cells as well as blood cells in various microfluidic channels. It could significantly speed up the process of microfluidic device design, which is usually time-consuming due to the large amount of trial-and-error experiments.
We will release the work by end of April and I will be happy to talk about more details by that time.
Tell me a bit about your GPU Computing background.
I started programming on the GeForce GTX 460 GPUs using OpenCL since 2010, and in 2012 I shifted entirely to CUDA C++.
Right now, I use mostly Kepler GPUs with high double-precision floating-point performance. I have been focused on accelerating particle-based simulations including All-Atom Molecular Dynamics (AAMD), Dissipative Particle Dynamics (DPD) and Smoothed Particle Hydrodynamics (SPH).
In fact, I have developed an entire GPU package (our USERMESO package), for the LAMMPS (Large-scale Atomic/Molecular Massively Parallel Simulator) particle simulator for DPD and SPH simulations. The package achieves 20x to 30x speed up on a single K20 GPU over 16 AMD CPU cores on a Cray XK7 compute node.
How has GPU computing impacted your research?
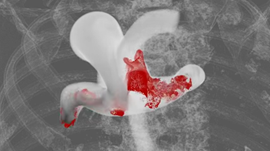
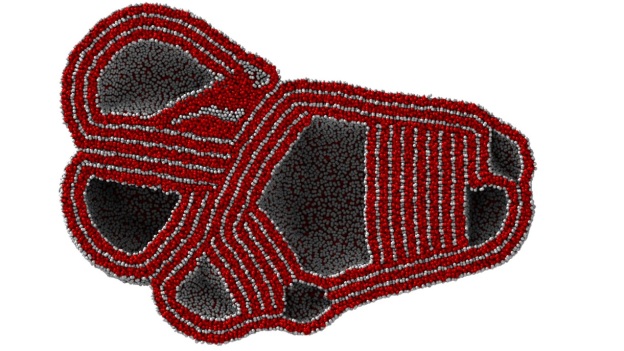
Our USERMESO package allows us to simulate DPD systems containing several millions of particles for millions of time steps on a daily basis during the study of the self-assembly behavior of amphiphilic polymers. The multi-compartment multi-walled vesicle, or simply think of it as a miniature cell, as Figure 1 shows, is only observable at a spatial-temporal scale that is tens of times larger, and tens of times longer than that covered by typical contemporary DPD simulations. With the USERMESO code we can perform such simulations daily with just 16 GPUs!
What approaches have you used to apply the CUDA platform to your research?
I use CUDA C++ and I design customized math routines for DPD simulations using hand-coded PTX assembly. The template metaprogramming feature of CUDA C++ also turns out to be handy for writing concise and efficient codes.
Which CUDA features and GPU libraries are you currently taking advantage of?
Streams, zero-copy memory, texture objects, PTX (parallel thread execution) assembly, warp-level vote/shuffle, template programming.
What new features of CUDA 7 have you used in your current project?
C++11. The benefit is two-fold:
- I can now directly compile device codes mixed with C++11 host codes. Our host-side C++11 code is a generic library that we use to concurrently couple the GPU-based particle solver with CPU-based continuum solvers. In earlier CUDA releases nvcc would fail if the host codes contain C++11 syntax, and simply passing –Xcompiler –std=c++11 to the host compiler cannot solve it. As a result I had to compile the host code separately and then link with device object files. With CUDA 7 I can simply compose everything in one file and compile by a single shot.
- It allows me to make partial specialization of devices functions, passing lambdas and set template default arguments, etc. This greatly improved my coding productivity.
Can you share some visual results?
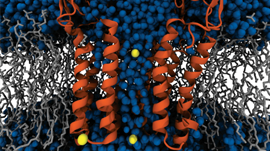
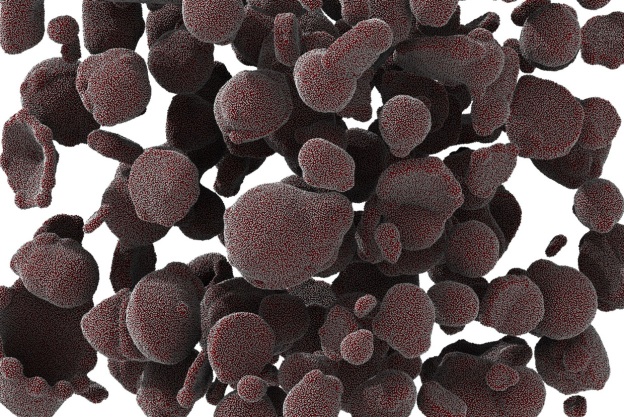
Figures 2 and 3 were from a system of vesicles spontaneously assembled from amphiphilic polymers in aqueous solution. The result, together with the USERMESO code and algorithm, is published in: Tang, Yu-Hang, and George Em Karniadakis. “Accelerating dissipative particle dynamics simulations on GPUs: Algorithms, Numerics and Applications.” Computer Physics Communications 185.11 (2014): 2809-2822.
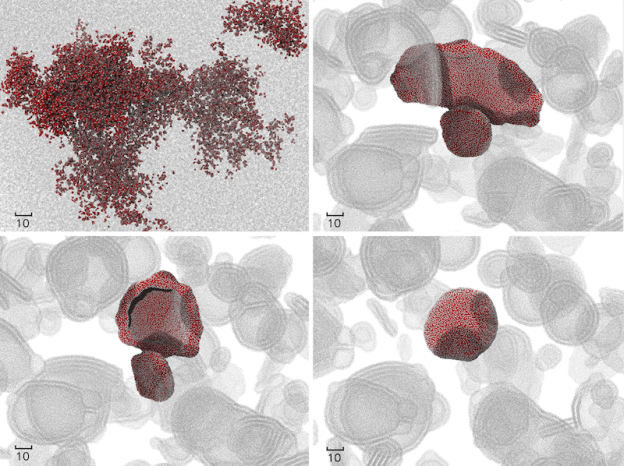
Can you share any of your algorithms and is USERMESO open source?
In USERMESO I invented a warp-synchronous neighbor list building algorithm that allows the neighbor list for a particle to be constructed deterministically in parallel by all the threads within a warp without using any atomic operations. This actually makes neighbor searching much faster than evaluating pairwise forces, while traditionally the searching takes longer time. For details and visualization of the algorithms you can check out my slides from the 2014 GPU Technology Conference.
And, yes our code USERMESO is open source; you can find it on my wiki page.
In terms of technology advances, what are you looking forward to in the next five years?
At the system level I look forward to GPUs that are more tightly coupled to other system parts like CPUs, RAM, interconnects and drives. In terms of the CUDA architecture I think configurable warp size and a faster and bigger non-coherent cache would benefit applications in both my area of interest and many other algorithms.
More About CUDA 7
- The Power of C++11 in CUDA 7
- CUDA 7 Release Candidate Feature Overview: C++11, New Libraries, and More
- C++11 in CUDA: Variadic Templates
Download CUDA 7 today! If you have tried it, please comment below and let us know your thoughts.