Dr. Joshua A. Anderson is a Research Area Specialist at the University of Michigan who was an early user of GPU computing technology. He began his career developing software on the first CUDA capable GPU and now runs simulations on one of the world’s most powerful supercomputers.
Dr. Joshua A. Anderson is a Research Area Specialist at the University of Michigan who was an early user of GPU computing technology. He began his career developing software on the first CUDA capable GPU and now runs simulations on one of the world’s most powerful supercomputers.
Anderson’s “contributions to the development and dissemination of the open source, GPU-enabled molecular simulation software, HOOMD-blue, which enables scientific computations with unprecedented speed” earned him the 2015 CoMSEF Young Investigator Award for Modeling and Simulation.
Brad Nemire: Can you talk a bit about your current research?
Joshua Anderson: I work with the Glotzer Group at the University of Michigan. We use computer simulation to discover the fundamental principles of how nanoscale systems of building blocks self-assemble, and to discover how to control the assembly process to engineer new materials. Specifically, we focus on the role of particle shape and how changing the shape can result in different material properties.
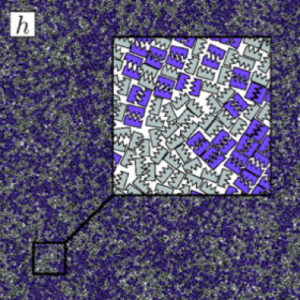
Over the past few years, I have been focusing on two-dimensional systems, using large scale simulations to study hexatic phase transitions for hard disks, and how patterning surfaces of polygons can create shape allophiles that improve self-assembly. The hexatic phase is an intermediate between the fluid and hexagonally ordered solid. In the hexatic phase, the orientation of bonds between particles has long range order, but translational order is short range and there is no crystal lattice. Shape allophiles are polygonal shapes cut so they fit together like puzzle pieces. These research projects are computationally demanding and could not have been run on any existing code. So before I could even begin the science research, I needed to develop, implement, and optimize the parallel algorithms necessary for these studies.
Hard particle Monte Carlo is a particle simulation technique similar to molecular dynamics. We use it to model nano- and colloidal-scale particles that have shape. Imagine a box full of tiny polyhedral dice shaken so that they are allowed to find their most favorable state. There are no attractive forces, simply a penalty force so that particles cannot overlap. Entropy alone causes systems of hard particles to self-assemble into a variety of complex ordered phases.
I implemented hard particle Monte Carlo as an addition to HOOMD-blue, an open source particle simulation toolkit that I created and of which I lead development. HOOMD-blue is highly configurable: users write Python job scripts that activate and configure features desired for the run. HOOMD-blue began as a molecular dynamics simulation engine. Here is an example job script that runs a Lennard-Jones fluid in molecular dynamics.
from hoomd_script import *
init.read_xml('init.xml')
lj = pair.lj(r_cut=2.5)
lj.pair_coeff.set('A', 'A', epsilon=1.0, sigma=1.0)
integrate.mode_standard(dt=0.005)
integrate.nvt(group=group.all(), T=1.2, tau=0.5)
run(1e5)
Using Python, users can easily create complex workflows: custom initialization routines, vary run parameters, analyze the state of the simulation and decide to continue or stop, or anything else the user can think of. The current v1.3 release of HOOMD performs molecular dynamics simulations with a variety of different force fields; the Monte Carlo simulation development I discuss here will be available in v2.0 soon.
BN: When and why did you start looking at using NVIDIA GPUs?
JA: Since middle and high school, I wanted to be a game developer and wrote my own graphics engines using a GeForce 256. But I was much more interested in physically realistic simulation engines behind the games, so it’s no surprise that I eventually become a computational physicist. Throughout college, I kept graphics programming as a hobby and even took some graduate-level CS courses on graphics. I started graduate school at the time when many were beginning to experiment with using graphics APIs to do computational research. I played around with some ideas for doing the same for molecular dynamics, and then the first CUDA beta was released. I was at a conference at the time, and I spent late nights at the hotel reading through the CUDA programming guide and planning out the initial data structures and parallel algorithms that would eventually become HOOMD-blue. At first, I had to convince my advisor, Alex Travesset, that it was worth buying a GeForce 8800 GTX (G80) for research—as I had not yet upgraded my own gaming PC. I compiled a test benchmark and e-mailed it to my brother, who did have a G80. The initial performance results were more then enough to convince my advisor. As a small research group, we didn’t have many funds for computational resources and had to squeeze in jobs when we could on the department’s small cluster. I wanted to write my own molecular dynamics simulation code so that I could run simulations on my workstation without needing to wait in the cluster queue.
BN: What is your GPU Computing experience and how have GPUs impacted your research?
JA: I started working with GPU computing in the early days of CUDA, using a single GPU in a workstation to develop code and run simulations. Now, I am part of a large research group with allocations on the Oak Ridge National Laboratory Titan and National Center for Supercomputing Blue Waters supercomputers. My current research project would not have been possible without the use of large GPU supercomputers. Using 4000 GPUs at a time on Titan, I can run simulations at dozens of different state points on 64 GPUs each and complete the entire set of runs in a few days. Using CPU-only clusters, or even smaller GPU clusters, the same batch of runs would have taken several months.
On the software development side of my work, the impact of GPUs is huge. HOOMD-blue wouldn’t exist without GPUs, as it was built from the ground up with GPU support. Most users come to the code because of the level of performance it provides on GPUs.
BN: Can you share some performance results?
In my most recent work on hard particle Monte Carlo, we benchmarked HOOMD-blue running a system of 16.7 million pentagons on Titan. At the strong scaling limit on 2048 GPUs (only 8192 particles per GPU), it could complete 10 million sweeps (one trial move per particle) in just 1.4 hours. However, there is no other code to compare to for these types of simulations, on GPU or CPU. For me, it is sufficient to say that some of the science done using HOOMD-blue would not have been possible without GPUs.
BN: How are you using GPUs?
JA: We write CUDA C++ kernels for most calculations in HOOMD-blue. Particle simulations do not lend themselves to library calls. For molecular dynamics, though, the implementation is fairly simple. In molecular dynamics, we need to evaluate forces on all particles and then integrate them forward in time according to the equations of motion. These operations are inherently parallel as they occur separately for each particle in the system. Early versions of HOOMD simply ran one thread per particle in CUDA kernels, limiting parallelism to N threads. In the strong scaling limit, however, the GPU becomes the bottleneck when N is small enough that there are not enough threads to fill the device. We expand these routines to run more than one thread per particle and use warp-synchronous stream compaction and reduction operations within a group of threads that cooperatively process each particle. We auto-tune the block size and number of threads per particle at run time to find the fastest performing launch parameters.
Hard particle Monte Carlo is not as simple as molecular dynamics. The formulation of Monte Carlo is to propose a trial move to one particle at a time, check for overlaps with the surrounding particles and accepts the move if there are no overlaps. This inherently serial operation needs to be extended to allow for parallelism. We use a checkerboard grid to propose trial moves in parallel in many cells without conflicts. This limits parallelism to fewer threads than particles, as only a subset of particles can be moved in parallel. The parallel algorithm must not only run fast, but it must also produce the correct results from a statistical mechanical standpoint. We performed tests with the hard disk system and found that a combination of shuffling the order of selected trial moves and rejecting moves that cross cell boundaries is necessary to match existing data.
BN: What NVIDIA technologies are you using?
JA: We primarily use hand-written CUDA C++ kernels. In a few cases we use Thrust or CUB library calls (sort, prefix sum, scan). Within our CUDA kernels, we take advantage of many Kepler-specific features when available: including shuffle() for warp-synchronous scan and reduction operations, and __ldg() for fast semi-random memory access and to avoid texture binding launch overheads.
For multi-GPU simulations, we have an alternate code path that uses GPUDirect-RDMA, but this is usually slower than hand-tuned code on current hardware.
BN: What are some of the biggest challenges in this project?
JA: The biggest challenge I needed to solve for the hard particle Monte Carlo code was avoiding divergence overheads. Detecting overlaps between two arbitrarily shaped polyhedra is computationally expensive, so threads spend a long time in this check. Each trial move has only a 20% chance of success, and once a failed trial move is detected no further processing needs to be done. With only one thread per trial move, this means that, on average, 80% of a warp (~25 threads) exits early and only ~7 threads are in the critical path and determine how long the warp runs. With some suggestions from NVIDIA Developer Technology Engineers, I experimented with many queue-scheduling algorithms to avoid this problem. After many attempts, I finally found one that reduced the kernel run time significantly. The final version of the kernel runs multiple threads per trial move, which perform early rejection tests on the nearby particles. Tests that fail the early rejection test are added into a block-wide work queue. Once the queue is full, each thread in the block processes one expensive overlap check (which may be for a trial move in a different thread) and puts the result in shared memory. In this way, on the 2nd queue-filling pass, threads that already know they have a failed trial move do not add any work to the queue and thus we get some of the early exit benefit. At the same time, the most expensive part of the computation is always done by all threads in the block at once, so divergence is minimized.

BN: What’s next? What’s your vision?
JA: HOOMD-blue continues to grow with more features added every month. The core simulation engine is complete and fully optimized for current generation GPUs. I am now spending much of my time working with contributors to the project to add specific ancillary features that enable more types of research to be done with the code. We are at a stage now where most research projects are not limited by the time it takes to run the simulations; rather, the bottleneck is the human time spent in configuring runs, managing data files, and analyzing results. I hope to return to my graphics programming roots and develop a Python-driven visualization engine for visualizing simulations run in HOOMD-blue—GPU-accelerated, of course, using NVIDIA OptiX. A visualization tool with the performance and flexibility of HOOMD-blue will reduce the time a researcher needs to spend analyzing data.
BN: In terms of technology advances, what are you looking forward to in the next five years?
JA: Most kernels in HOOMD-blue are bound by memory bandwidth. I look forward to the high-bandwidth memory available with Pascal. Explicit memory management that we already use in HOOMD-blue will always be faster, but I do look forward to trying automatic Unified Memory transfers in less performance-critical data analysis code. Less time spent implementing the analysis means more time to do research.
BN: What impact do you think your research will have on this scientific domain? Why?
JA: HOOMD-blue has a large impact in its domain. Even as a relatively new project, more than 122 peer-reviewed publications that use it have been published, growing faster every year. As a modern, high-performance, general-purpose particle toolkit, it appeals to young researchers. It also has strong GPU support, and is easy to configure and use on any system.
BN: Where do you obtain technical information related to NVIDIA GPUs and CUDA?
JA: I always go to the NVIDIA CUDA Programming Guide. That was where I learned CUDA, when there was no other information, and that is where I go back to. The companion Best Practices and Tuning Guides are also very useful. The NVIDIA Developer Forums are also a great resource. At one time, I frequently answered users questions there, but am now unfortunately too busy with other responsibilities. There are still many CUDA experts and NVIDIA engineers that quickly answer technical questions on the forums.